Description
The Redscar© Database is a collection of wound images. The images were obtained from the clinical trial https://clinicaltrials.gov/ct2/show/NCT05485233
This image database contains photographs of abdominal surgery wounds. The images contain metadata in their name, with the following structure
{image_id}_infection=INF_capture=CAP_resolution=RES.png
Where:
-{image_id} is a string of 25 characters, including lowercase and uppercase letters and numbers.
-INF is a boolean, representing if the image has been classified as infected (1) or without infection (0).
-CAP is an integer, representing the phase of the clinical study in which the image was captured.
-RES is a boolean, representing if the image has a good resolution (1), more than 500x500, and (0) otherwise.
For each image, there are three additional manually constructed images containing useful information about the objects present. Specifically:
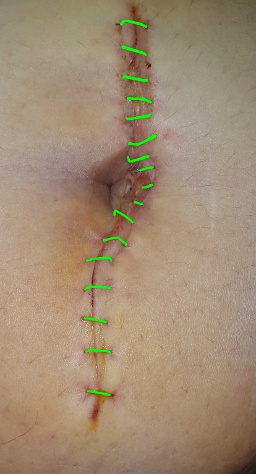
-IMAGES: Folder containing the original images, without any modification.
-GT_WOUND_MASK: Folder containing binary images, representing the mask with the position of the wound.
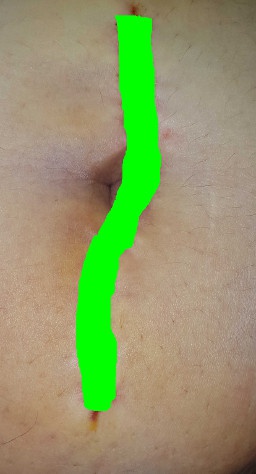
-GT_WOUND_COLORMASK: Folder containing colour images, representing the mask with the position of the wound drawn in green on the original
image.
-GT_STAPLES_COLORMASK: Under construction.
Each of these folders contains the image with the same name, so that it is easy to identify them in each category. In the following table, there are some statistics about the dataset taking "infection" and "resolution" as main categorigal variables:
| Infection | |||
| 0 | 1 | ||
|
Valid |
0 | 159 (40.25%) | 5 (1.27%) |
| 1 | 207 (52.53%) | 23 (5.84%) | |
Samples
Please see below some examples of the images that are part of the database.
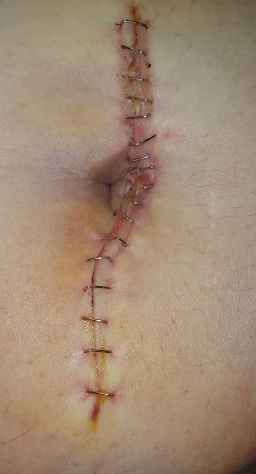
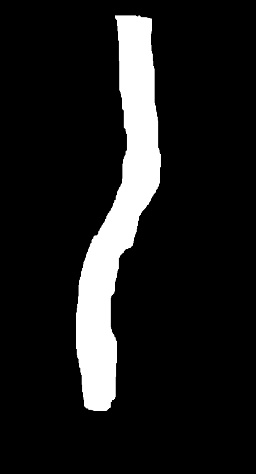
| IMAGES | GT_WOUND_MASK |
| GT_WOUND_COLORMASK | GT_STAPLES_COLORMASK |
Access
Access to the database is restricted for privacy reasons and to account for how much it is used, and in which projects. To get free access to the database, please contact us by email:
with the subject: "Request Redscar© database access".
Acknowledgements
This work was partially supported by the R+D+i Project PID2020-113870GB-I00-“Desarrollo de herramientas de Soft Computing para la Ayuda al Diagnóstico Clínico y a la Gestión de Emergencias (HESOCODICE)”, funded by MCIN/AEI/10.13039/501100011033/.

 OpenDemo
OpenDemo